Load and prepare data
## Registered S3 method overwritten by 'quantmod':
## method from
## as.zoo.data.frame zoo
library(mlr3)
library(ggplot2)
library(forecast)
# National Centers for Environmental Information, National Oceanic and Atmospheric Administration
muc = read.csv("https://www.ncei.noaa.gov/data/global-summary-of-the-year/access/GM000004199.csv")
nyc = read.csv("https://www.ncei.noaa.gov/data/global-summary-of-the-year/access/USW00094728.csv")
muc = muc[, c("DATE", "PRCP", "TAVG", "TMIN", "TMAX")]
nyc = nyc[, c("DATE", "PRCP", "TAVG", "TMIN", "TMAX")]
nyc[is.na(nyc$TAVG), ]
## DATE PRCP TAVG TMIN TMAX
## 1 1869 1149.9 NA NA NA
## 152 2020 NA NA NA NA
# remove first year
nyc = nyc[-1, ]
muc_start <- muc$DATE[1]
muc_end <- muc$DATE[nrow(muc)]
nyc_start <- nyc$DATE[1]
nyc_end <- nyc$DATE[nrow(nyc)]
# nyc check missing
all_years = nyc_start:nyc_end
missing_years = setdiff(all_years, nyc$DATE)
# add missing years
all_years = muc_start:muc_end
missing_years = setdiff(all_years, muc$DATE)
missing_years_df = data.frame(DATE = missing_years, PRCP = NA, TAVG = NA, TMIN = NA, TMAX = NA)
muc = rbind(muc, missing_years_df)
muc = muc[order(muc$DATE), ]
head(muc)
## DATE PRCP TAVG TMIN TMAX
## 1 1879 864.7 6.37 2.10 10.64
## 2 1880 1172.7 8.24 3.96 12.52
## 3 1881 813.5 7.69 3.32 12.06
## 4 1882 982.6 8.22 3.80 12.63
## 5 1883 887.4 7.78 3.63 11.94
## 6 1884 801.9 8.55 4.23 12.87
# keep information on station , temp and precipitation
muc = muc[, c("PRCP", "TAVG", "TMIN", "TMAX")]
nyc = nyc[, c("PRCP", "TAVG", "TMIN", "TMAX")]
muc_ts = ts(muc, start = muc_start, end = muc_end, frequency = 1)
nyc_ts = ts(nyc, start = nyc_start, end = nyc_end, frequency = 1)
Generate Task
task = TaskRegrForecast$new(
id = "nyc",
backend = nyc_ts,
target = c("TAVG", "TMIN", "TMAX"))
task$print()
## <TaskRegrForecast:nyc> (151 x 4)
## * Target: TAVG, TMIN, TMAX
## * Properties: multivariate
## * Features (1):
## - dbl (1): PRCP
Learner
learner = LearnerRegrForecastVAR$new()
learner$train(task, row_ids = 1:140)
learner$model
##
## VAR Estimation Results:
## =======================
##
## Estimated coefficients for equation TAVG:
## =========================================
## Call:
## TAVG = TAVG.l1 + TMIN.l1 + TMAX.l1 + const + PRCP
##
## TAVG.l1 TMIN.l1 TMAX.l1 const PRCP
## 4.0771421122 -1.8509164028 -1.7015792696 4.7167644305 0.0003791629
##
##
## Estimated coefficients for equation TMIN:
## =========================================
## Call:
## TMIN = TAVG.l1 + TMIN.l1 + TMAX.l1 + const + PRCP
##
## TAVG.l1 TMIN.l1 TMAX.l1 const PRCP
## 14.9032339288 -6.9015015629 -7.4926644859 3.6618636553 0.0005266401
##
##
## Estimated coefficients for equation TMAX:
## =========================================
## Call:
## TMAX = TAVG.l1 + TMIN.l1 + TMAX.l1 + const + PRCP
##
## TAVG.l1 TMIN.l1 TMAX.l1 const PRCP
## -6.681064783 3.165766819 4.054795062 5.780513431 0.000233528
Predict
p = learner$predict(task, row_ids = 141:150)
p$response
## row_id TAVG TMIN TMAX
## 1: 141 12.18238 8.278473 16.08544
## 2: 142 12.39421 8.536479 16.25221
## 3: 143 12.16620 8.204436 16.12647
## 4: 144 12.14030 8.144487 16.13512
## 5: 145 12.20270 8.207059 16.19768
## 6: 146 12.11141 8.065036 16.15647
## 7: 147 12.08404 8.009921 16.15695
## 8: 148 12.10114 8.016975 16.18420
## 9: 149 12.30878 8.292957 16.32444
## 10: 150 12.28554 8.264854 16.30535
## [1] NA NA NA NA NA NA NA NA NA NA
Rolling Window CV
rr = rsmp("RollingWindowCV", fixed_window = F)
rr$instantiate(task)
resample = resample(task, learner, rr, store_models = TRUE)
resample$predictions()[1:2]
## [[1]]
## <PredictionForecast> for 5 observations:
## row_id truth.TAVG truth.TMIN truth.TMAX response.TAVG response.TMIN
## 18 10.52 6.43 14.61 10.74273 6.744645
## 19 9.69 5.70 13.67 11.04004 7.109768
## 20 11.55 7.76 15.34 11.22040 7.303086
## 21 11.64 7.76 15.51 10.97845 7.021135
## 22 12.30 8.45 16.15 10.79115 6.823432
## response.TMAX
## 14.73874
## 14.97048
## 15.13860
## 14.93531
## 14.75750
##
## [[2]]
## <PredictionForecast> for 5 observations:
## row_id truth.TAVG truth.TMIN truth.TMAX response.TAVG response.TMIN
## 20 11.55 7.76 15.34 10.63703 6.792219
## 21 11.64 7.76 15.51 10.84419 6.956840
## 22 12.30 8.45 16.15 10.83183 6.899231
## 23 11.23 7.45 15.00 10.78568 6.828165
## 24 10.39 6.56 14.23 10.76442 6.822249
## response.TMAX
## 14.48044
## 14.73034
## 14.76355
## 14.74211
## 14.70496
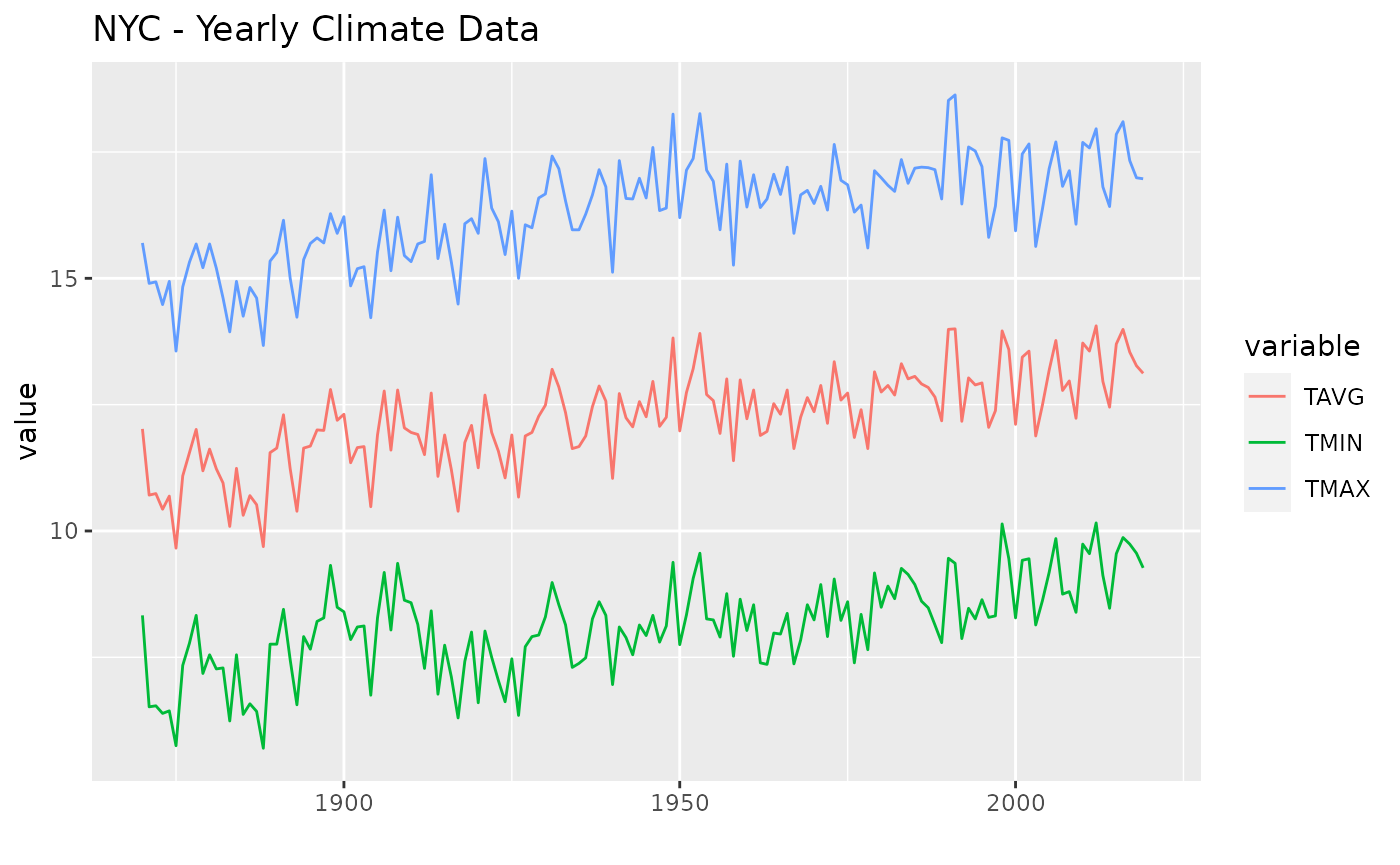
Plotting
## Warning: Removed 3 row(s) containing missing values (geom_path).
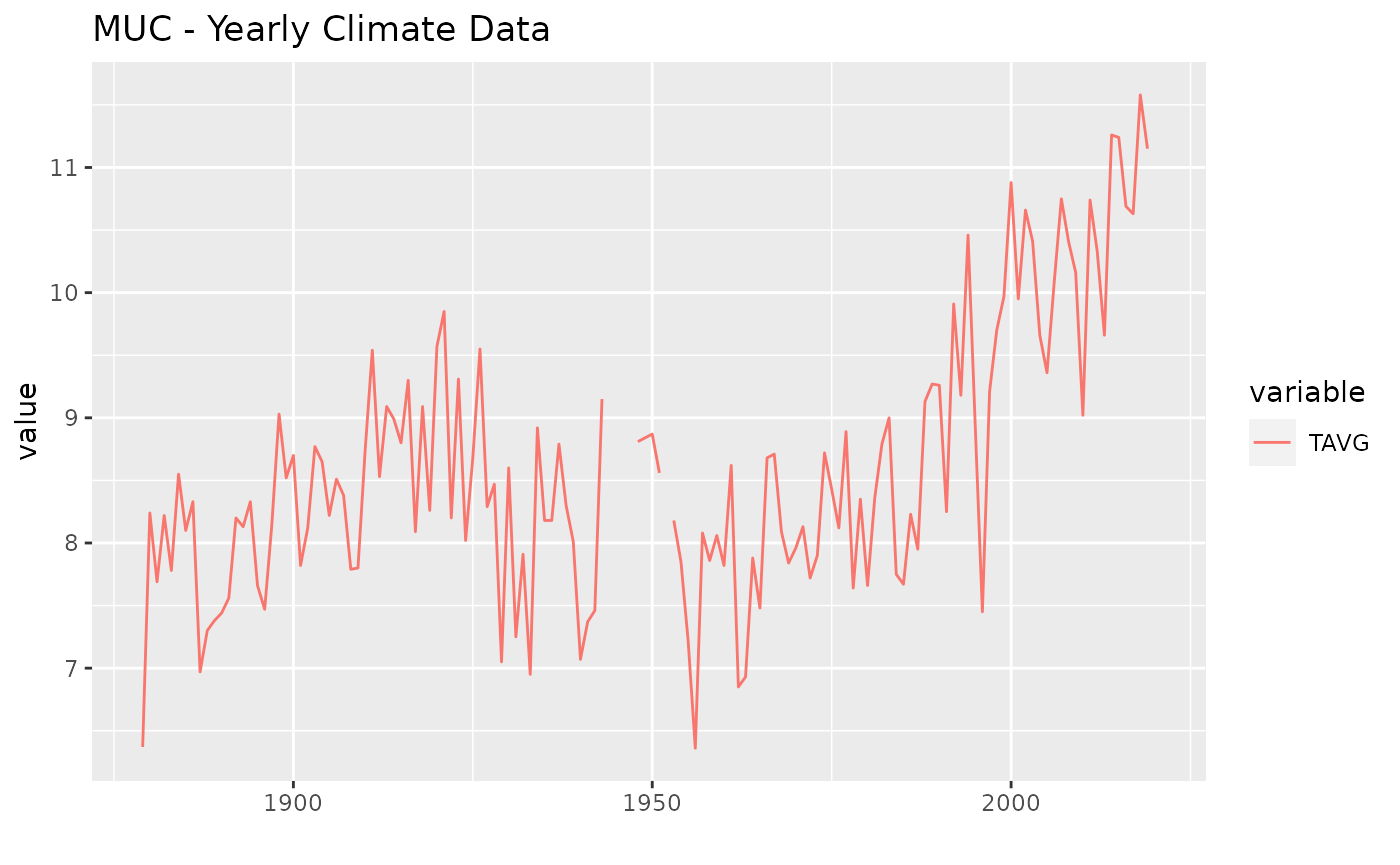
task = TaskRegrForecast$new(
id = "muc",
backend = ts(muc[c("TAVG", "PRCP")], start = muc_start, end = muc_end, frequency = 1),
target = "TAVG")
autoplot(task) + ggtitle("MUC - Yearly Climate Data")
## Warning: Removed 1 row(s) containing missing values (geom_path).
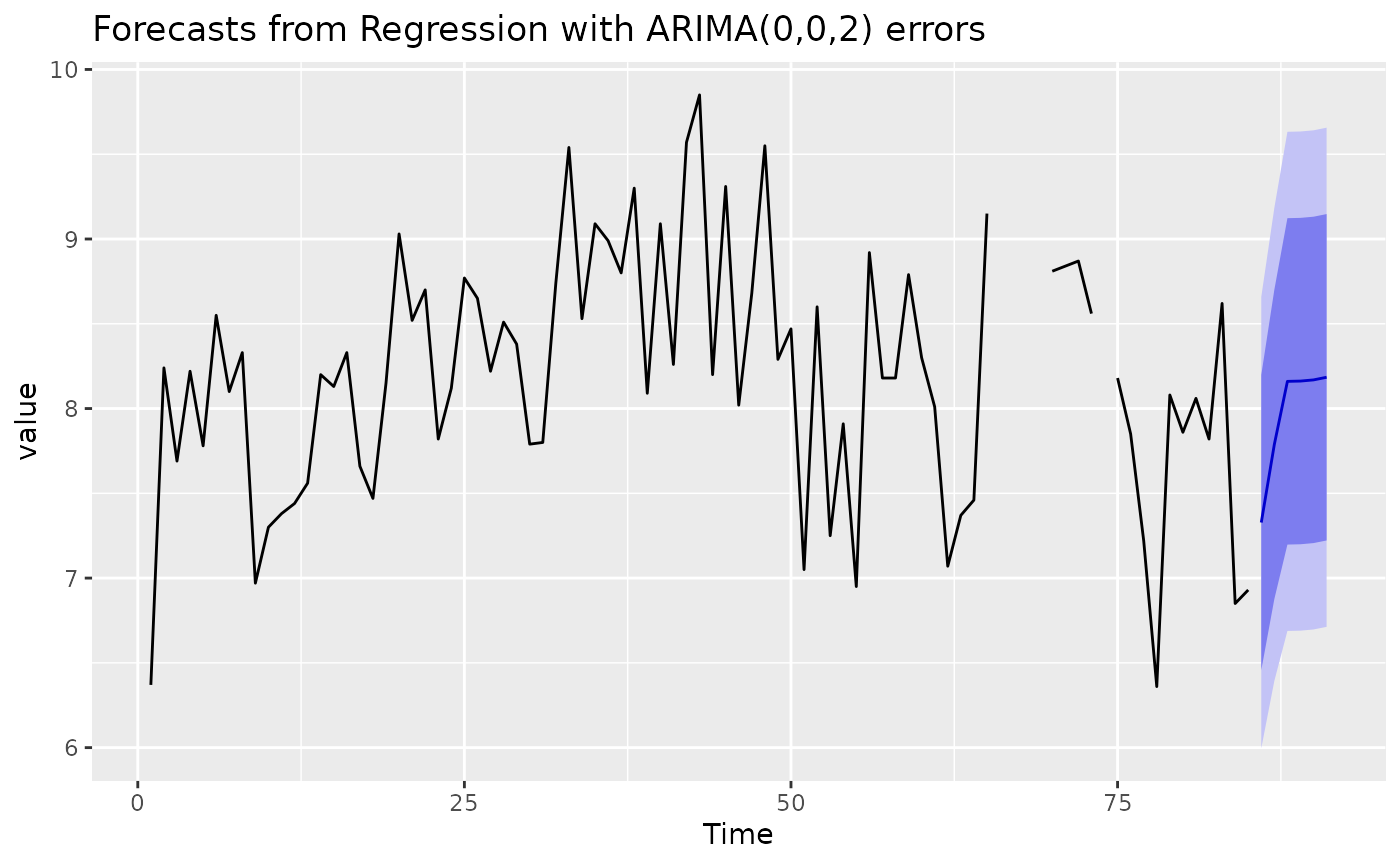
learner = LearnerRegrForecastAutoArima$new()
learner$train(task, row_ids = 1:85)
learner$model
## Series: task$data(rows = task$row_ids, cols = task$target_names)
## Regression with ARIMA(0,0,2) errors
##
## Coefficients:
## ma1 ma2 intercept PRCP
## 0.3129 0.3529 8.3595 -2e-04
## s.e. 0.1208 0.1113 0.5113 5e-04
##
## sigma^2 estimated as 0.4615: log likelihood=-80.94
## AIC=171.88 AICc=172.64 BIC=184.1
p = learner$predict(task, row_ids = 86:136)
p$se
## [1] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA
## [26] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA
## [51] NA
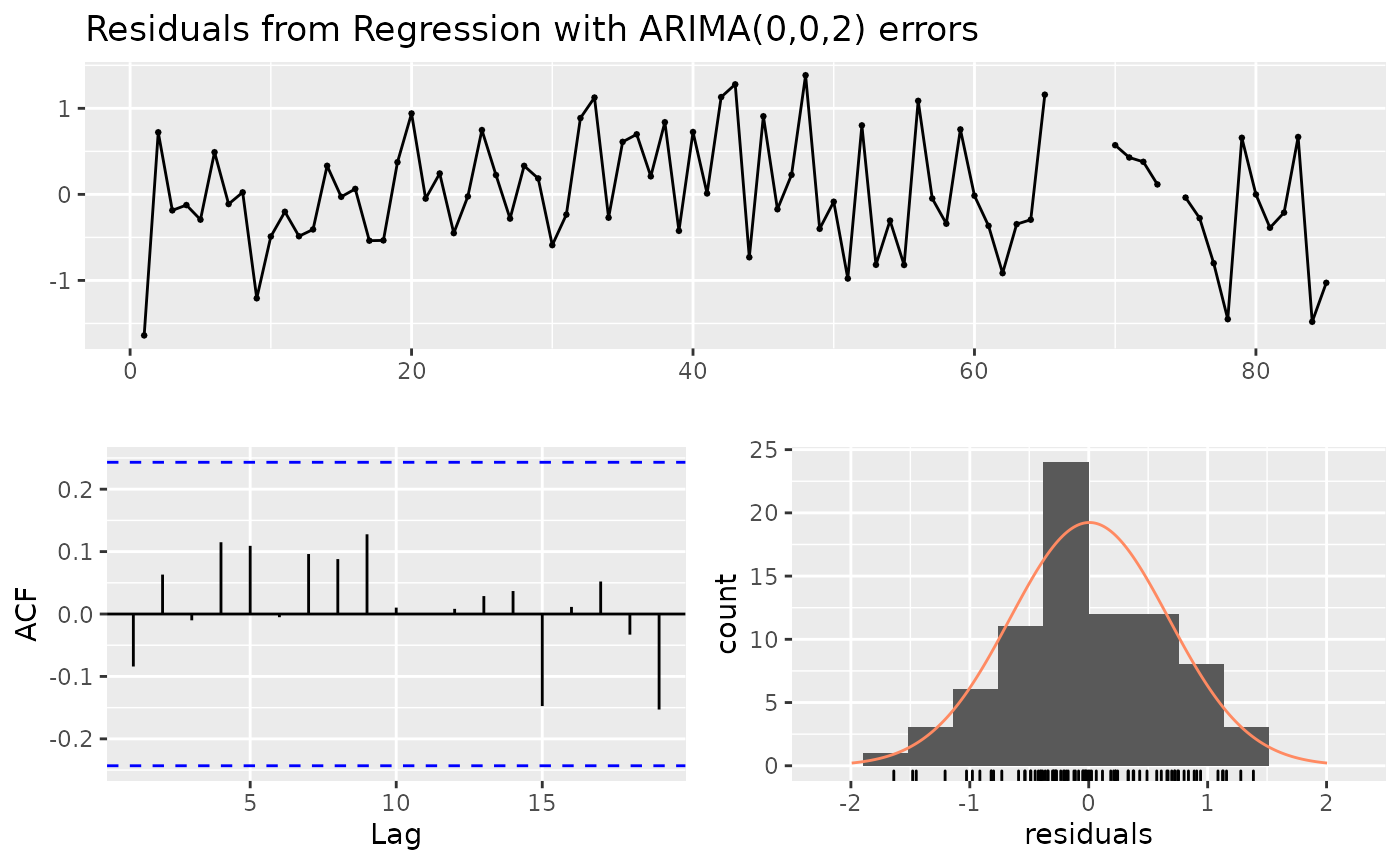
##
## Ljung-Box test
##
## data: Residuals from Regression with ARIMA(0,0,2) errors
## Q* = 4.9348, df = 6, p-value = 0.5522
##
## Model df: 4. Total lags used: 10